A new study sheds light on the development of Scandinavian settlement from the late Neolithic period to modern times. The research aims to evaluate the genomic results partly also from the perspectives of linguistics and archaeology. The research is so extensive that getting familiar with it takes a lot of time, so I won't even try to draw conclusions here. A few points already at this point. The term East Scandinavian created in the study mainly means the area of Sweden. Its counterweight is southern Scandinavia, mainly meaning the Danish region. The mixing of these areas into modern Scandinavians took place 3000-4000 years ago in southern Scandinavia. On the basis of this, it must be assumed that the migration took place from north to south, which was also brought up in the previous study dealing with the issue. From the point of view of linguistics, the interesting question of where the Scandinavian and Germanic language group was born still remains open, even though the issue is sidestepped cross-disciplinaryly by means of genetics. No more than Eastern Sweden and the Western Baltic can be reached here, and no evidence is presented. From the point of view of Finland, two samples from Ostrobothnia are interestingly found in the new material, which at a quick look look more like Finns, in contrast to previous Sami and Scandinavian type samples from the same area. In itself, the fact that all these genotypes can be found in the same area in Ostrobothnia is not surprising, because the area was the starting point of the eastern trade route of the Scandinavians before it moved to the Gulf of Finland and the Daugava river.
Kalevan ja Untamon geenit
Monday, March 18, 2024
Monday, February 19, 2024
Razib Khan's special interest in Finnish culture/ancestry
Razib Khan is a Pakistani-American genetist being active in population genetics and has a long work history in creating tools for consumer business in that area. I have followed his online writings and impact in creating software tools for companies selling genetic ancestry analyses. All the time he has had a special interest in Finnish culture and genetic history. It has been pleasure to read his wonderful visions about us and see the progress in his comprehension. Read and enjoy about his latest writings.
https://www.razibkhan.com/p/finnish-brains-baiting-and-bottlenecks
https://www.razibkhan.com/p/weirdness-as-a-national-pastime
https://www.razibkhan.com/p/go-west-young-siberian
https://www.razibkhan.com/p/from-deepest-siberia-to-europes-edge
https://www.razibkhan.com/p/frontier-finns-cabins-rakes-and-indians
Razib Khans blog: https://www.gnxp.com/
Wednesday, January 3, 2024
Nothing new in Sweden since the Bronze Age?
Thanks to Danish researchers for releasing Scandinavian Bronze Age samples, available at ENA. I downloaded bam files of those Scandinavian Bronze Age I1 men mentioned in my previous post, processed them and ran a PCA plot, including present-day Europeans. Here is a screenshot of the study preprint picture of corresponding samples:
The plot (below) shows evidently that all those I1 samples locate in the same position with present-day Swedes, excluding Swedish individuals showing Finnish influence. The similarity with Swedes after 3000-4000 years actually a bit surprises me. Deeper eigenvectors show a bit more western tendency, though. Pity I have no modern Danish samples to compare with them. Finnish I1 samples don't differ in any aspect from the modern Finnish population, which is an expected result and shows identical impact of long-term local influence as it has been the case also in Sweden, see the preprint picture where the BA I1 sample group diverges from the ancient Central European sample group.
Wednesday, December 20, 2023
Preprint Allentoft et al. 2024
The preprint of the study estimates the age of the I1 male population of Southern Scandinavia to be around 4000 years and defines the direction of their arrival as northeast. The time period coincides with the late Scandinavian Neolithic (Scandinavian Dagger Period) and the early Scandinavian Bronze Age. The newcomers are connected to the stone cist burial, a new type of burial common in southern Scandinavia. The burial method in question was common in Gotland as well, and the oldest graves in this area in Estonia are associated with western influences and migration from the west (interestingly, the Gotland saga, Gutasaga, mentions a migration to an island in Estonia, probably to Saaremaa and also to Poland. Then the Danish prehistorical tradition tells about elite groups who migrated from the north, warred against jutes and beat them. They formed the root of Danes). Later, the Scandinavian influence decreased in Estonia and a new burial method, the Tarand burial, took its place.
Today's oldest I1 clades based on living human samples are 4600 years old. The oldest I1 samples based on ancient DNA are 7000-7500 years old and were found in Gotland (Stora Förvar 11) and Hungary (BAB5). The Hungarian sample exclusively represents the early European Neolithic population, while the Gotland sample represents mainly local hunter-gatherers, but with a piquant European Neolithic addition. As a side note, Sweden's most common clade, L22, is 3900 years old according to the Yfull website, the main group of Finnish I1 men is 2300 years old and its upstream group is 2800 years old. Regarding to the Estonian I1 we know only the size of proportion, because so far the research has shed light only on clades of Estonian haplotypes N1 and R1a. The proportion of Estonian I1 is 15% according to earlier studies.
Stone cist graves in Gotland
Stone cist graves in Estonia
Estonian Tarand graves, usually connected to the Baltic Finnic migration from the east. Tarand graves followed the stone cist period.
In Finland, Bronze Age graves are common, there are thousands of discovered Bronze Age graves and stone cairns assumed to be graves, but I could not find maps specific to the grave type. After reading several archaeological studies related to stone cist burials, I am still unsure of their origin and historical chronology. One of the reasons for this may be that archaeologists did basic research in both Finland and Sweden before the digital revolution, and these studies are not available online.
Wednesday, December 6, 2023
Finnish ydna samples on the autosomal map
In his own abstract, A. Pruessner presents the geographical distribution of the autosomal inheritance of Finnish ydna. I have been waiting for academic research on this topic for a long time. What is being said now about the division of the N haplogroup into eastern and southwestern groups has been known for more than 10 years based on Ftdna's project data, so the presentation does not bring much new information to those interested in the matter. Pruessner's abstract does not say much about the geography of other haplogroups. Of course, for example I1 forms geographical clusters, but mainly from a common Finnish root. On the other hand, the origin of the N1 clusters refers to different angles of entry into the Finnish area. Inspired by Pruessner's presentation, I decided to do a PCA analysis using Finnish samples from the 1000genones project as data. The data includes 38 male samples out of a total of 99 samples. Due to the brevity of the ydna data in the material, I cut the definition to two characters (N1, I1, R1, one I2 removed), even if a more precise result would have been possible in some of the samples. I also did the same test with the narrower Eurogenes data. As a final conclusion, excluding ouliers in R1 and N1, two in both, it can be stated that the autosomal distribution of all yDna groups seems to be more or less similar in the entire Finnish population, although the narrower Eurogenes data points I1 towards Scandinavia and R1 towards Eastern Europe.
Figures:
1 Pruessner's abstract
2 PCA based on Eurogenes global data
3 PCA based on 1000genomes, West Eurasian view
Friday, November 17, 2023
North Europe using Eurogenes G25-coordinates
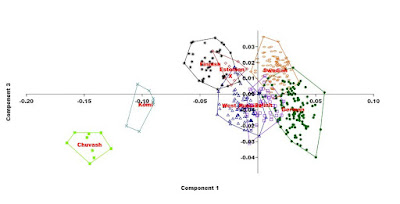
Too often in studies common heritability and genetic drift are mixed up in a population context. Pretty much research has been ruined this way. This inevitably happens when comparing the heritage of local populations. The Eurogenes coordinate system compares samples in the global coordinate system and eliminates local drift, and by using Eurogenes G25 coordinates, an objective picture of the common inheritance of population groups can be obtained even at the local sample level without the error caused by local genetic drift.
Following plots are made using the original Eurogenes G25 data. The vertical axis on the three dimensional picture (sticks) shows similarty between Swedish, Finnish and Estonian samples. East European samples with Siberian admixture (Komi and Chuvash) are taken to the plot to point out the Siberian admixture, gaining the maximum on the PC 1.
Tuesday, October 31, 2023
Thoughts about the past and the present
Policy warning. My readers may have noticed that I don't like the enthusiasm with which Finnish genetic research is looking for Finns' ethnic roots in Russia. Unlike language, which has only one root, the genetic heritage is multi-rooted. Genetically, the inhabitants of Finland are a mixture of the southern (Estonia) and eastern (Karelia, Vepsä) Baltic Finno-Ugrians, Scandinavians of different time periods, Sami and increasingly other ethnic groups. These groups, except for the last 30 years of immigration, have been intermingling for hundreds of years. This is forgotten, as was also the case in the Estonian study, which I discussed in my previous post. Geneticists in Finland and Estonia are ideologically very single-minded in their desire to find a multidisciplinary solution by simplifying things. Of course, it is necessary to find out where our language comes from, and linguistics studies that.
Things get more complicated when the Russian view of Finns' roots is added to the previous one. I am unusually well aware that, according to the Russian point of view, the Finno-Ugric peoples living in Russia are called Finns, and Finns as one of them. In reality, the linguistic and ancient genetic roots of the Baltic Finns formed a reasonably homogenous entity that differed from other Finno-Ugric people. Baltic Finnic speakers diverged from other Finno-Ugric language speakers already 1500 years before the Russia as a state was created and formed their own genetic pool. The political problems between Russia and the West will last, and the ideology of Finnish researchers about the origin of Finns in Russia may weaken the status of the Finno-Ugric people of Russia in their own homeland. We should see the difference between the past and the present, we should not create historical illusions that create problems in the development of communities.